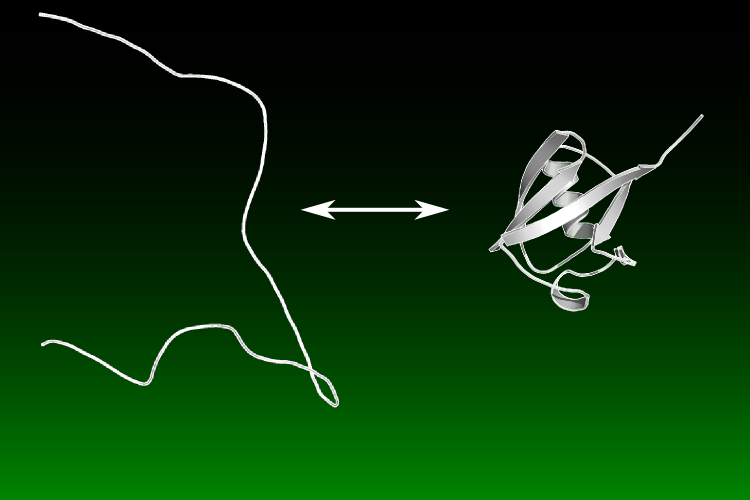
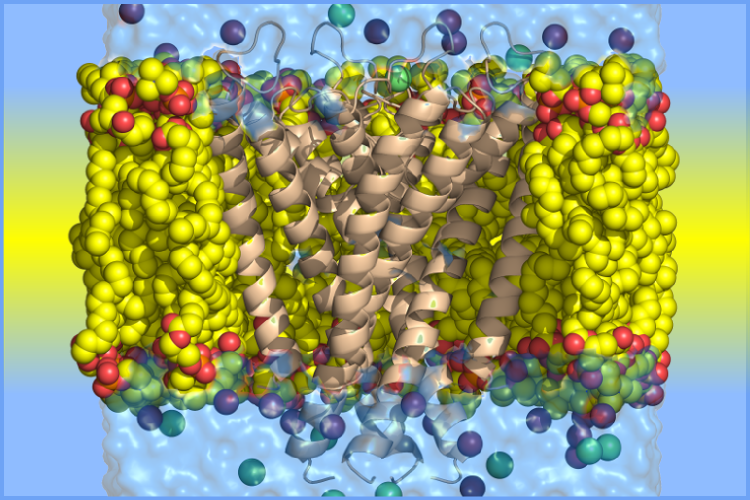
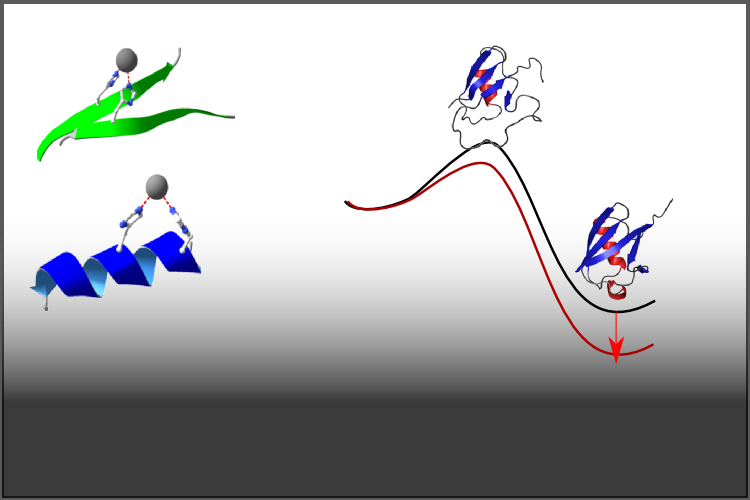
Research Areas in The Sosnick Group at The University of Chicago
Recent Publications
M.C. Baxa, X. Lin, C.D. Mukinay, S. Chakravarthy, J.R. Sacheleben, S. Antilla, N. Hartrampf, J.A. Riback, I.A. Gagnon, B.L. Pentelute, P.L. Clark, T.R. Sosnick, "How hydrophobicity, side chains, and salt affect the dimensions of disordered proteins" Prot Sci 33(5):e4986 (2024).
H. Glauninger, J.A.M. Bard, C.W. Hickernell, E.M. Airoldi, W. Li, R.H. Singer, S. Paul, J. Fei, T.R. Sosnick, E.W.J. Wallace, D.A. Drummond, "Transcriptome-wide mRNA condensation precedes stress granule formation and excludes stress-induced transcripts" bioRxiv 2024.04.15.589678
S.K. Kik, D. Christopher, H. Glauninger, C.W. Hickernell, J.A.M. Bard, M. Ford, T.R. Sosnick, D.A. Drummond, "An adaptive biomolecular condensation response is conserved across environmentally divergent species" Nat Comm 15, 3127 (2024).
X. Lin, P. Haller, N. Bavi, N. Faruk, E. Perozo, T.R. Sosnick, "Folding of Prestin’s Anion-Binding Site and the Mechanism of Outer Hair Cell Electromotility" eLife 12:RP89635
T.R. Sosnick, "AlphaFold developers Demis Hassabis and John Jumper share the 2023 Albert Lasker Basic Medical Research Award" J Clin Invest. 133 (19) (2023).