Conformational Entropy
Conformational Entropy
The change in entropy upon folding is a significant thermodynamic parameter. The conformational entropy is one component that is intimately related with solvent entropy, and cannot be decoupled experimentally without making assumptions.
Our studies have identified that the loss of conformational entropy is not as large as previously thought and is dominated by the loss of backbone entropy. These calculations have been integrated into a server for predicting the change in conformational entropy using a native PDB structure as input.
Approach
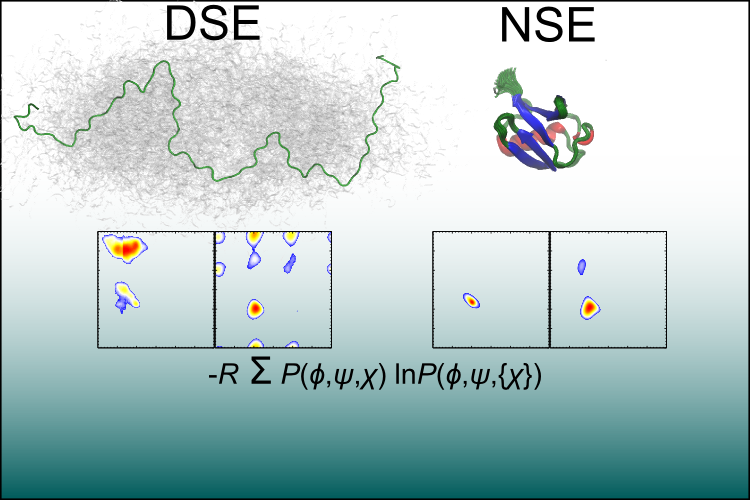
- We utilized a denatured state ensemble built using a statistical potential as a starting point for computing the conformational entropy of the denatured state using all-atom molecular dynamics.
- Native MD of the PDB structure provides the entropy of the native state.
- Backbone and sidechain dihedral angles distributions are generated for each residue and single and joint Shannon entropies are calculated.
- Predicting entropy loss from an input structure can be found at server listed below
Links:
PLOPS Predict the loss of conformational entropy upon folding